Algorithms for Characterizing Copy Number Variation in Human Genome
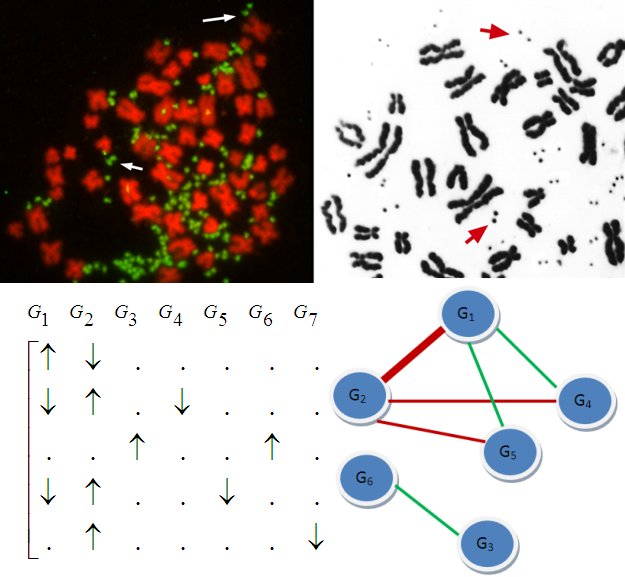
Not long ago, it was discovered that individuals may differ in copy
numbers of their genes,
meaning that a segment of DNA may have more or less copies than usual
in an individual's chromosome. Recent research suggests that
these variations are associated with many diseases including Autism
and Schizophrenia. Copy number
variation (CNV) in somatic cells also underly various cancers. Copy
numbers are usually identified using SNP microarrays, however,
short-read sequence data is emerging as an important resource
for characterizing structural variation in human genome.
As an interdisciplinary group of researchers at
Case
Western Reserve University, we develop algorithms for fast and accurate identification
of rare and de novo CNVs, as well as copy number polymorphisms (CNPs) and
other forms of genomic variation (e.g., loss of heterozygosity) from these two data sources. We
further extend these algorithms to identify small indels with applications to error correction
in next generation sequencing, fine tuning of the alignment of short reads
to the reference human genome, and characterization of tissue heterogeneity.
With a view to enabling personalized genomics applications,
we apply these algorithms to the identification of copy number variants, as well as
single nucleotide polymorphisms, genes, genetic interactions, pathways, and networks
that are associated with complex diseases.
Software
- LOQUM: A logistic regression
based algorithm for recalibrating the mapping quality of short-read sequence
alignments.
- SEAL: A comprehensive short
read sequencing simulation and alignment tool evaluation suite implemented
in Java.
- COKGEN:
An R package for optimization-based identification of rare and de novo
copy number variants from SNP microarray data.
Publications
- M. Ruffalo, M. Koyuturk, S. Ray, and T. LaFramboise.
Accurate estimation of short read mapping quality for next
generation genome sequencing, Bioinformatics Suppl. on 11th
European Conference on Computational Biology (ECCB),
in press.
- S. Erten, M. Ayati, Y. Liu, M. R. Chance, and M. Koyuturk.
Algorithms for detecting complementary SNPs within a region of
interest that are associated with diseases,
3rd ACM Conf. Bioinformatics, Computational Biology
and Biomedicine (ACM-BCB'12), in press.
- Y. Liu, S. Maxwell, T. Feng, X. Zhu, R. C. Elston, M. Koyuturk, and M. R. Chance.
Gene, pathway and network frameworks to identify epistatic interactions of
single nucleotide polymorphisms derived from GWAS data. BMC Systems Biology,
in press.
- G. Bebek, M. Koyuturk, N. D. Price, and M. R. Chance.
Network
biology methods integrating biological data for translational scie\
nce,
Briefings in Bioinformatics, 13(4): 446-459, 2012.
- G. Nickel, J. Barnholtz-Sloan, M. P. Gould, S. McMahon, A. Cohen,
M. D. Adams, K. Guda, A. E. Sloan, and T. LaFramboise.
Characterizing mutational heterogeneity in a glioblastoma patient with
double recurrence.
PLoS One, 7(4): e35262, 2012.
- Y.Liu, M. Koyuturk, J. Barnholtz-Sloan, and M. R. Chance.
Gene interaction
enrichment and network analysis to identtfy dysregulated pathways in cancer,
BMC Systems Biology, 6:65, 2012.
- M. Ruffalo, T. LaFramboise, and M. Koyuturk.
Comparative analysis of algorithms for next generation sequencing read alignment.
Bioinformatics, 27(20): 2790-2796, 2011.
- K. Wilkins and T. LaFramboise. Losing balance: Hardy-Weinberg disequilibrium as a marker
for recurrent loss-of-heterozygosity in cancer.
Human Molecular Genetics, 20(24):4831-4839, 2011.
- G. Yavas, M. Koyuturk, and T. LaFramboise.
Optimization algorithms for identification and genotyping of copy number polymorphisms
in human populations. 5th IAPR Int'l Conf. on Pattern
Recognition in Bioinformatics (PRIB'10), 74-85, 2010.
- G. Yavas, M. Koyuturk, M. Ozsoyoglu, M. P. Gould, and T. LaFramboise.
COKGEN:
A software for the identification of rare copy number variation from SNP microarrays.
Pacific
Symposium on Biocomputing (PSB'10), 371-382, 2010.
- G. Yavas, M. Koyuturk, M. Ozsoyoglu, M. P. Gould, and T. LaFramboise.
An optimization
framework for unsupervised identification of rare copy number variation from SNP array
data.
Genome Biology, 10:R119, 2009.
People

Katie Wilkins
Undergraduate Student, Computer Science/Biochemistry
(Now Ph.D. student at Cornell University)

Matthew Ruffalo
Ph.D. Student, Computer Science

Daniel Savel
Ph.D. Student, Computer Science

Marzieh Ayati
Ph.D. Student, Computer Science

Gokhan Yavas
Ph.D. Student, Computer Science
(Now post-doctoral fellow at Case Comprehensive Cancer Center)

Associate Professor,
Genetics & Genome Sciences

Associate Professor,
Electrical Engineering & Computer Science
Acknowledgments
This project is supported
by National Science Foundation
Award IIS-0916102.
| Not long ago, it was discovered that individuals may differ in copy numbers of their genes, meaning that a segment of DNA may have more or less copies than usual in an individual's chromosome. Recent research suggests that these variations are associated with many diseases including Autism and Schizophrenia. Copy number variation (CNV) in somatic cells also underly various cancers. Copy numbers are usually identified using SNP microarrays, however, short-read sequence data is emerging as an important resource for characterizing structural variation in human genome. As an interdisciplinary group of researchers at Case Western Reserve University, we develop algorithms for fast and accurate identification of rare and de novo CNVs, as well as copy number polymorphisms (CNPs) and other forms of genomic variation (e.g., loss of heterozygosity) from these two data sources. We further extend these algorithms to identify small indels with applications to error correction in next generation sequencing, fine tuning of the alignment of short reads to the reference human genome, and characterization of tissue heterogeneity. With a view to enabling personalized genomics applications, we apply these algorithms to the identification of copy number variants, as well as single nucleotide polymorphisms, genes, genetic interactions, pathways, and networks that are associated with complex diseases. |

|
Software
- LOQUM: A logistic regression based algorithm for recalibrating the mapping quality of short-read sequence alignments.
- SEAL: A comprehensive short read sequencing simulation and alignment tool evaluation suite implemented in Java.
- COKGEN: An R package for optimization-based identification of rare and de novo copy number variants from SNP microarray data.
Publications
- M. Ruffalo, M. Koyuturk, S. Ray, and T. LaFramboise. Accurate estimation of short read mapping quality for next generation genome sequencing, Bioinformatics Suppl. on 11th European Conference on Computational Biology (ECCB), in press.
- S. Erten, M. Ayati, Y. Liu, M. R. Chance, and M. Koyuturk. Algorithms for detecting complementary SNPs within a region of interest that are associated with diseases, 3rd ACM Conf. Bioinformatics, Computational Biology and Biomedicine (ACM-BCB'12), in press.
- Y. Liu, S. Maxwell, T. Feng, X. Zhu, R. C. Elston, M. Koyuturk, and M. R. Chance. Gene, pathway and network frameworks to identify epistatic interactions of single nucleotide polymorphisms derived from GWAS data. BMC Systems Biology, in press.
- G. Bebek, M. Koyuturk, N. D. Price, and M. R. Chance. Network biology methods integrating biological data for translational scie\ nce, Briefings in Bioinformatics, 13(4): 446-459, 2012.
- G. Nickel, J. Barnholtz-Sloan, M. P. Gould, S. McMahon, A. Cohen, M. D. Adams, K. Guda, A. E. Sloan, and T. LaFramboise. Characterizing mutational heterogeneity in a glioblastoma patient with double recurrence. PLoS One, 7(4): e35262, 2012.
- Y.Liu, M. Koyuturk, J. Barnholtz-Sloan, and M. R. Chance. Gene interaction enrichment and network analysis to identtfy dysregulated pathways in cancer, BMC Systems Biology, 6:65, 2012.
- M. Ruffalo, T. LaFramboise, and M. Koyuturk. Comparative analysis of algorithms for next generation sequencing read alignment. Bioinformatics, 27(20): 2790-2796, 2011.
- K. Wilkins and T. LaFramboise. Losing balance: Hardy-Weinberg disequilibrium as a marker for recurrent loss-of-heterozygosity in cancer. Human Molecular Genetics, 20(24):4831-4839, 2011.
- G. Yavas, M. Koyuturk, and T. LaFramboise. Optimization algorithms for identification and genotyping of copy number polymorphisms in human populations. 5th IAPR Int'l Conf. on Pattern Recognition in Bioinformatics (PRIB'10), 74-85, 2010.
- G. Yavas, M. Koyuturk, M. Ozsoyoglu, M. P. Gould, and T. LaFramboise. COKGEN: A software for the identification of rare copy number variation from SNP microarrays. Pacific Symposium on Biocomputing (PSB'10), 371-382, 2010.
- G. Yavas, M. Koyuturk, M. Ozsoyoglu, M. P. Gould, and T. LaFramboise. An optimization framework for unsupervised identification of rare copy number variation from SNP array data. Genome Biology, 10:R119, 2009.
People
 |
Katie Wilkins Undergraduate Student, Computer Science/Biochemistry |
 |
Matthew Ruffalo Ph.D. Student, Computer Science |
 |
Daniel Savel Ph.D. Student, Computer Science |
 |
Marzieh Ayati Ph.D. Student, Computer Science |
 |
Gokhan Yavas Ph.D. Student, Computer Science |
 |
Associate Professor, Genetics & Genome Sciences |
 |
Associate Professor, Electrical Engineering & Computer Science |